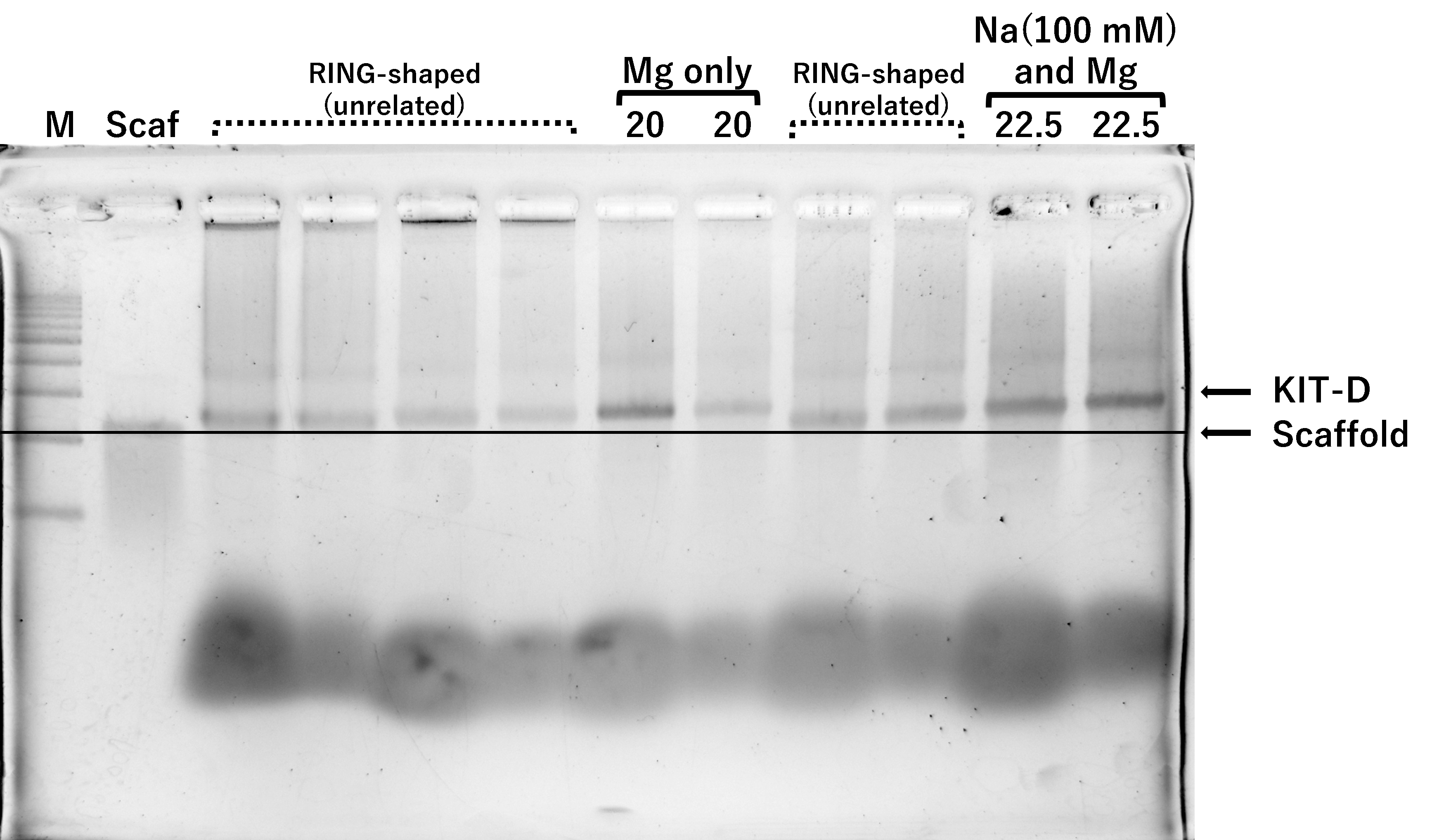
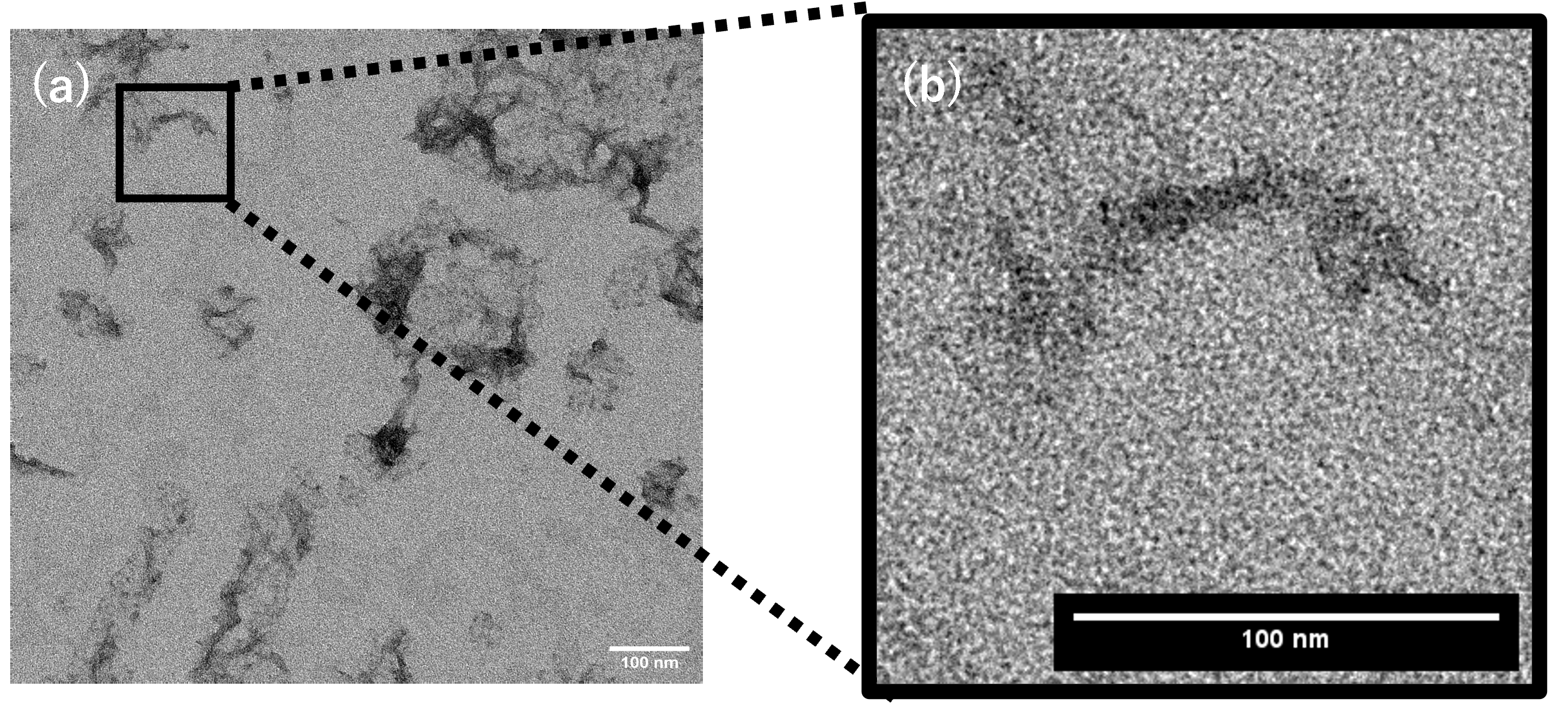
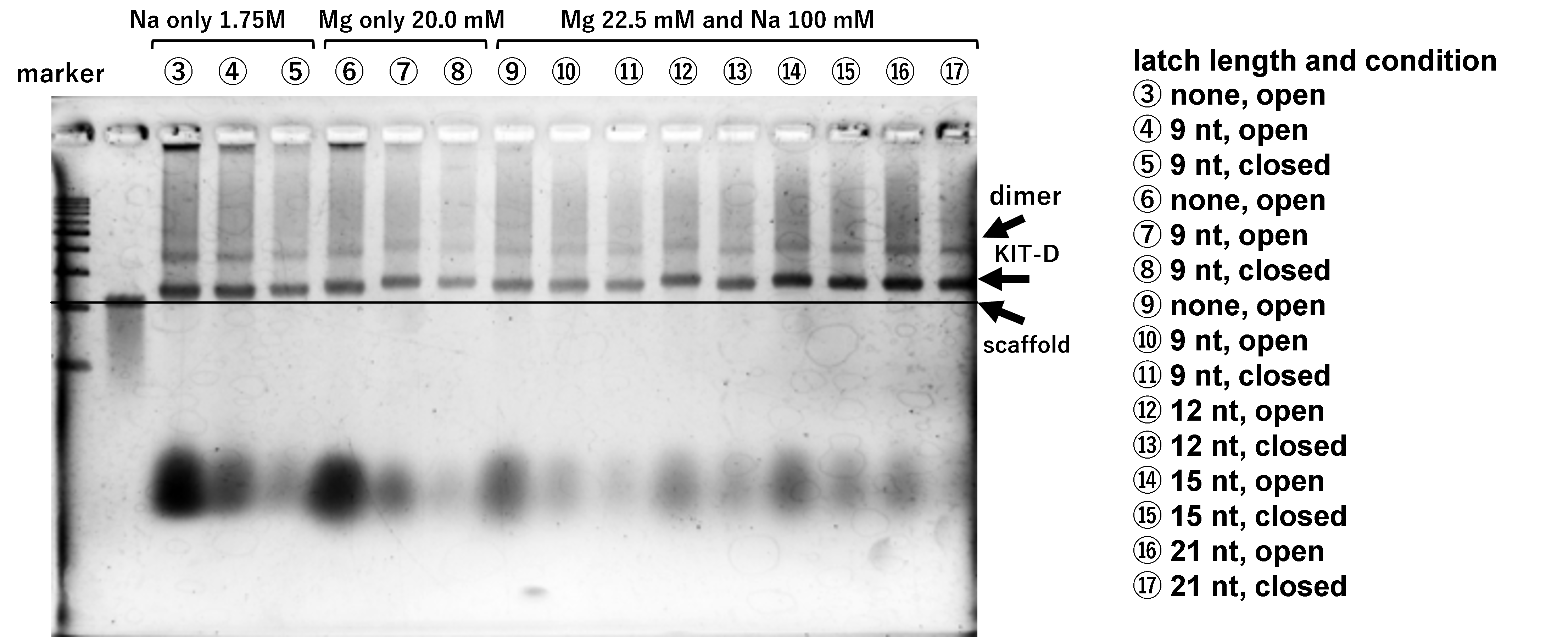
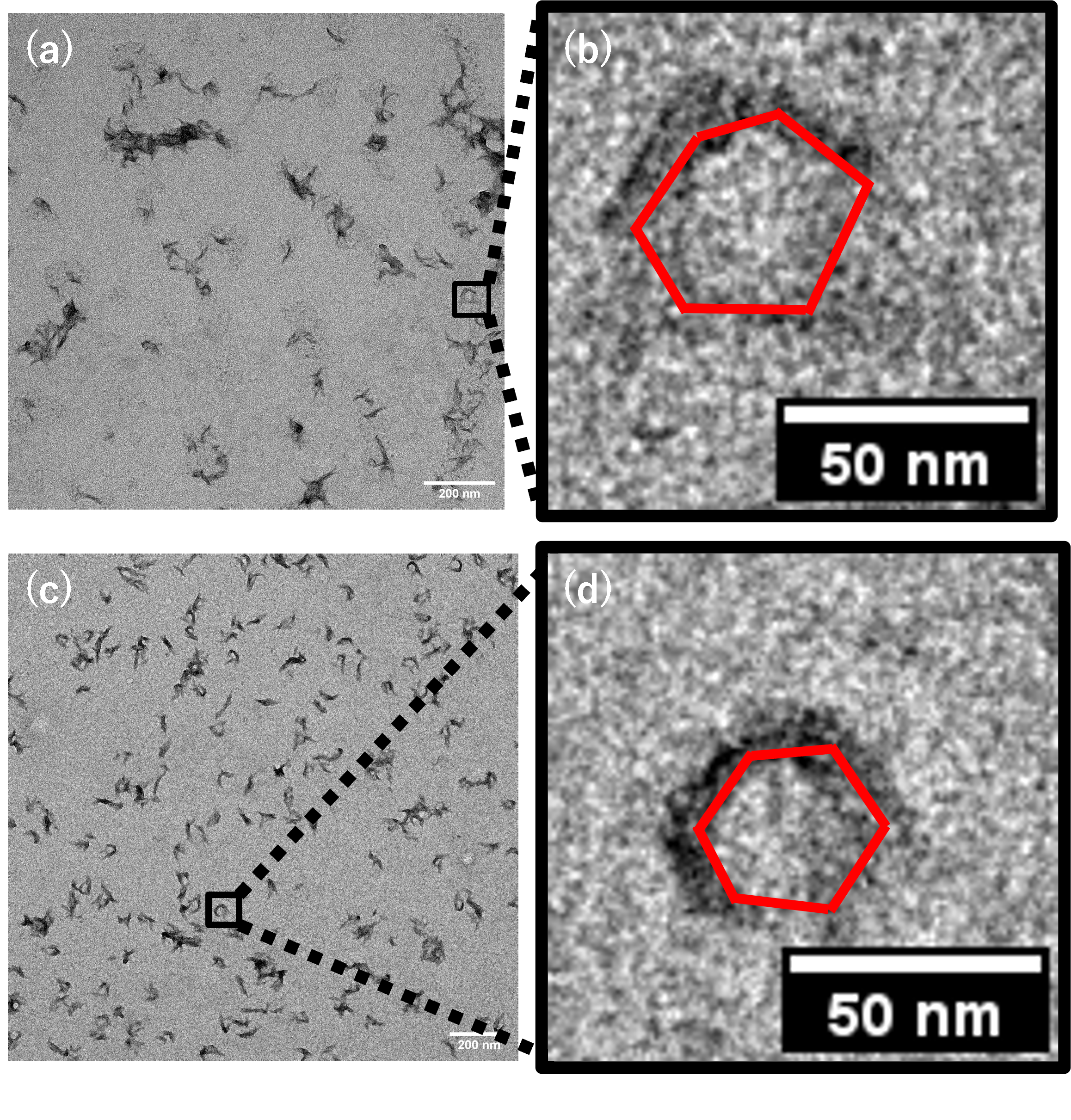
Optimization of structure formation conditions
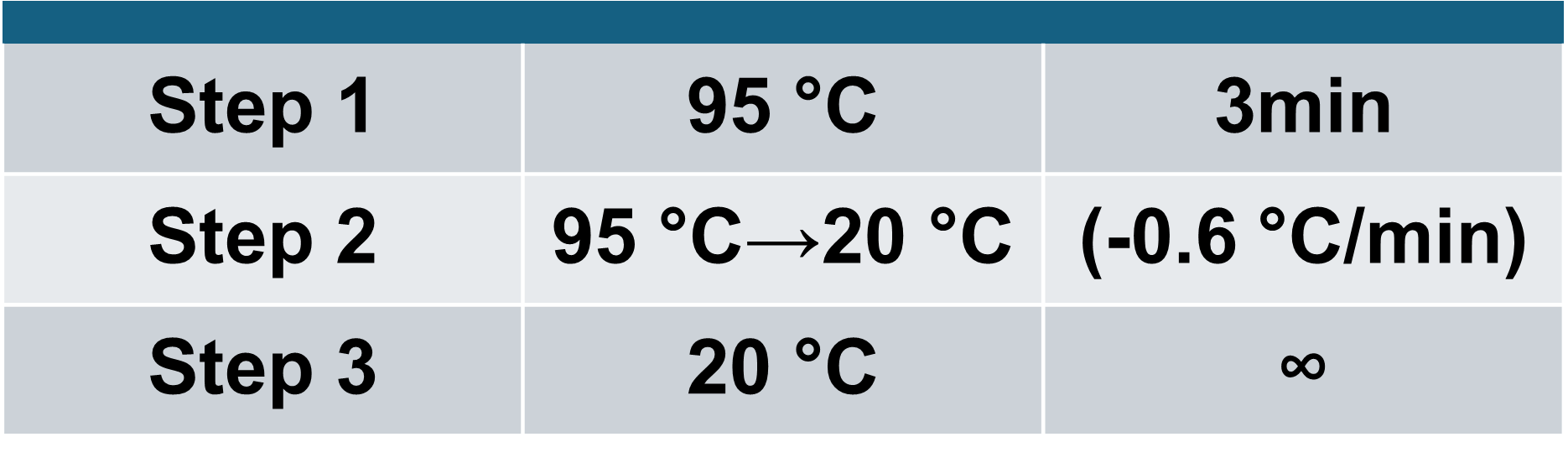
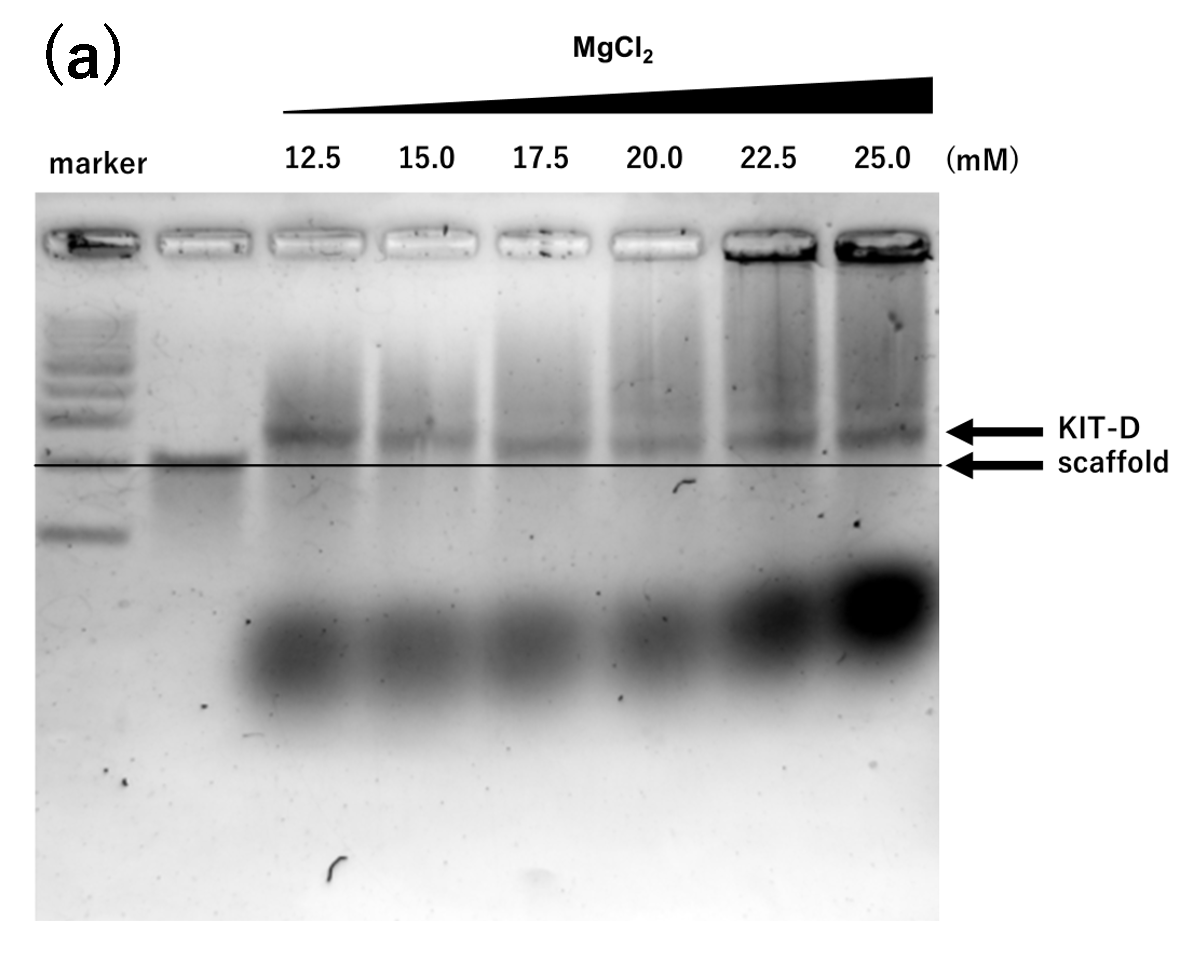
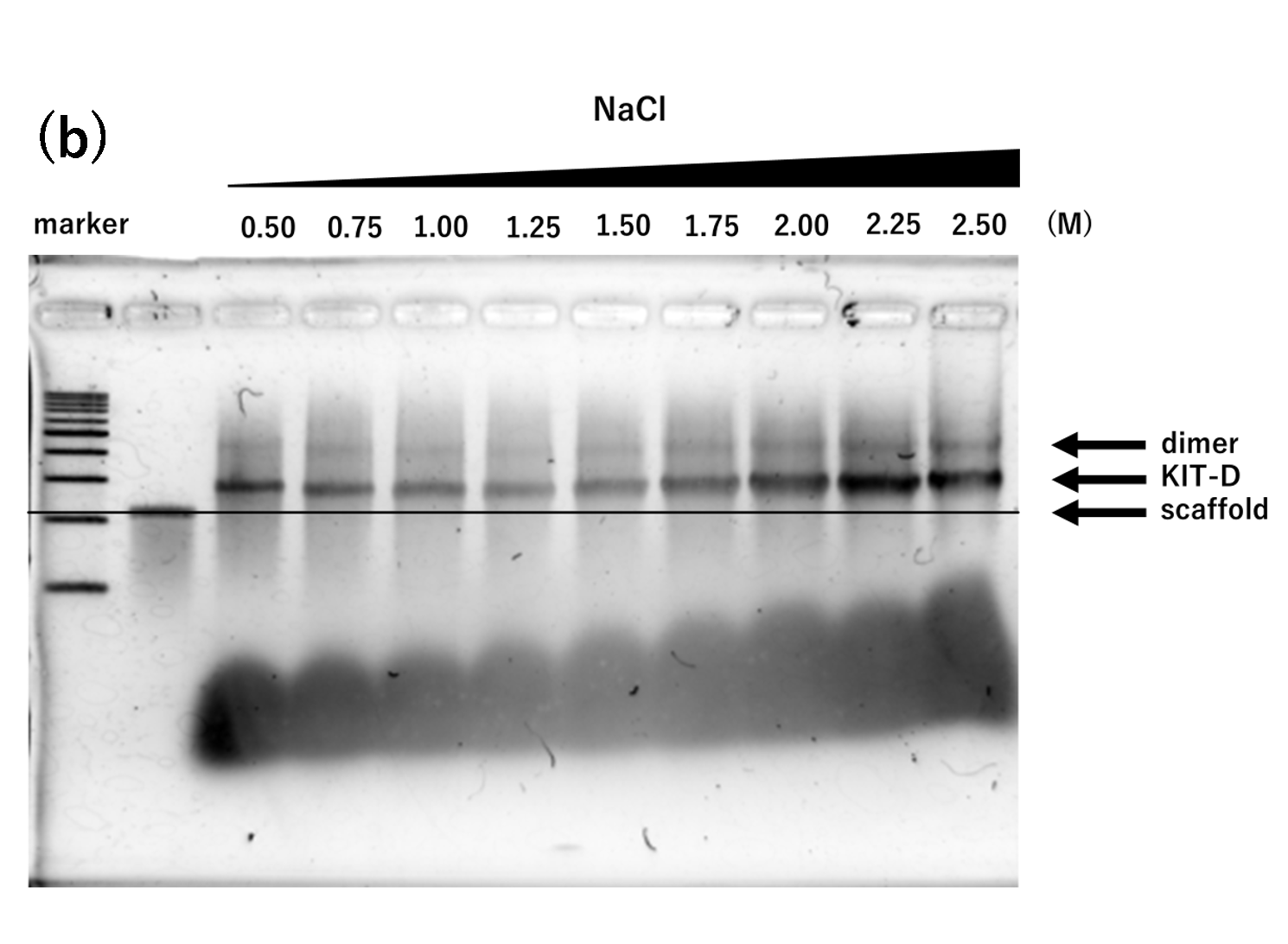
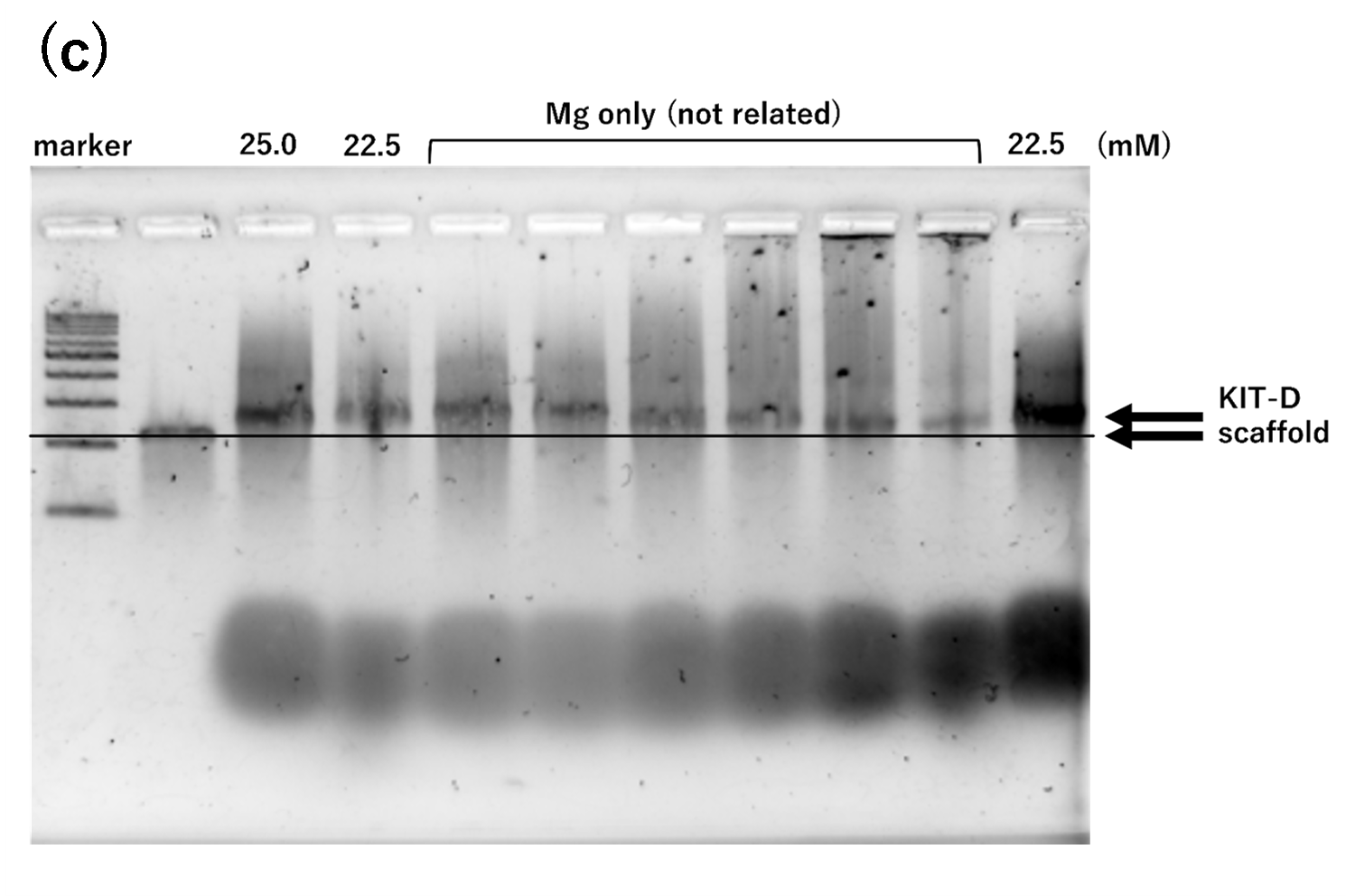
Figure 1. agarose gel electrophoresis of samples annealed for 2 hours
(a) Mg only, 2h, Lane 1: marker, Lane 2: 2 nM scaffold, Lane 3: 12.5
mM, Lane 4: 15.0 mM, Lane 5: 17.5 mM, Lane 6: 20.0 mM, Lane 7: 22.5
mM, Lane 8: 25.0 mM
(b) Na only, 2h, Lane 1: marker, Lane 2: 2
nM scaffold, Lane 3: 0.50 M, Lane 4: 0.75 M, Lane 5: 1.00 M, Lane 6:
1.25 M, Lane 7: 1.50 M, Lane 8: 1.75 M, Lane 9: 2.00 M, Lane 10:
2.25 M, Lane 11: 2.50 M
(c) Na(100 mM) and Mg, 2h, Lane 1:
marker, Lane 2: 2 nM scaffold, Lane 3: 25 mM, Lane 4: 22.5 mM, Lane
5~10: Mg only (unrelated), Lane 11: retry of Lane 4

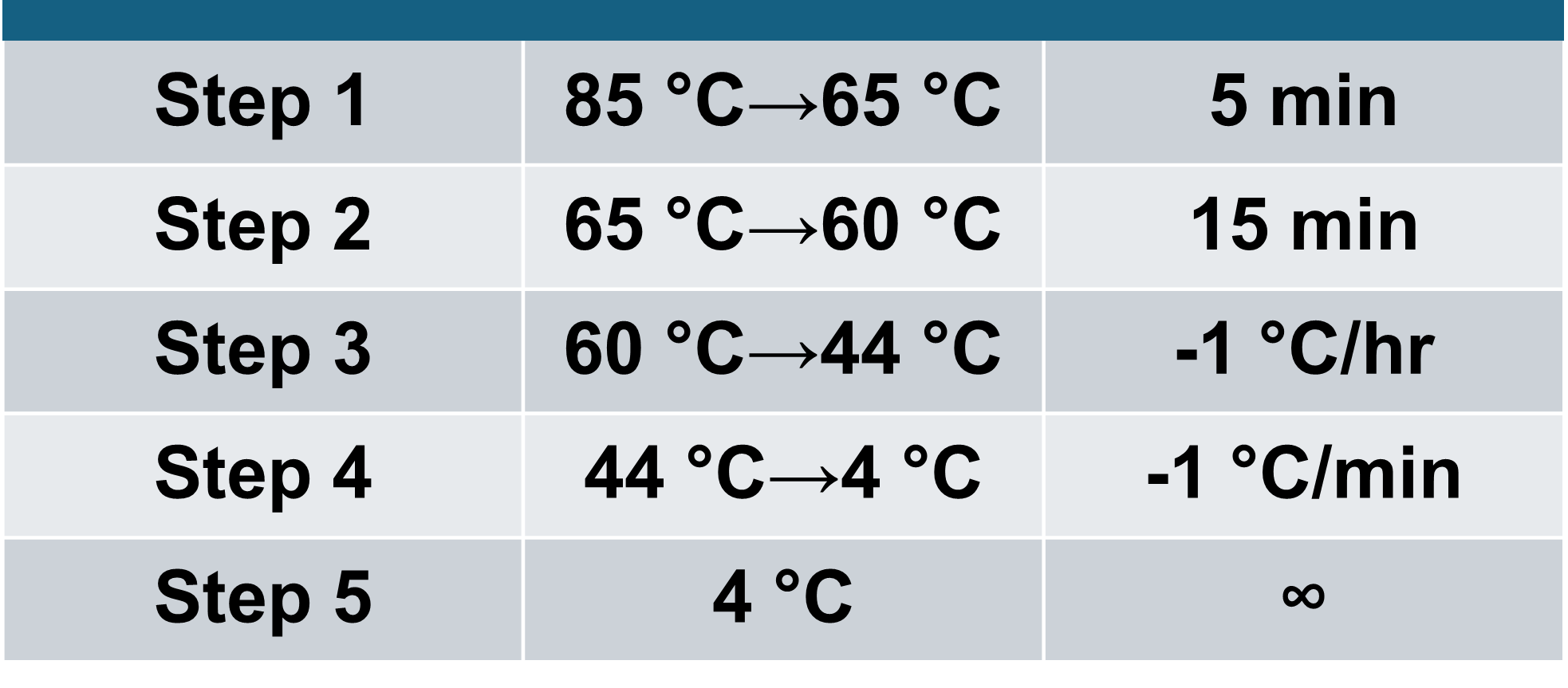
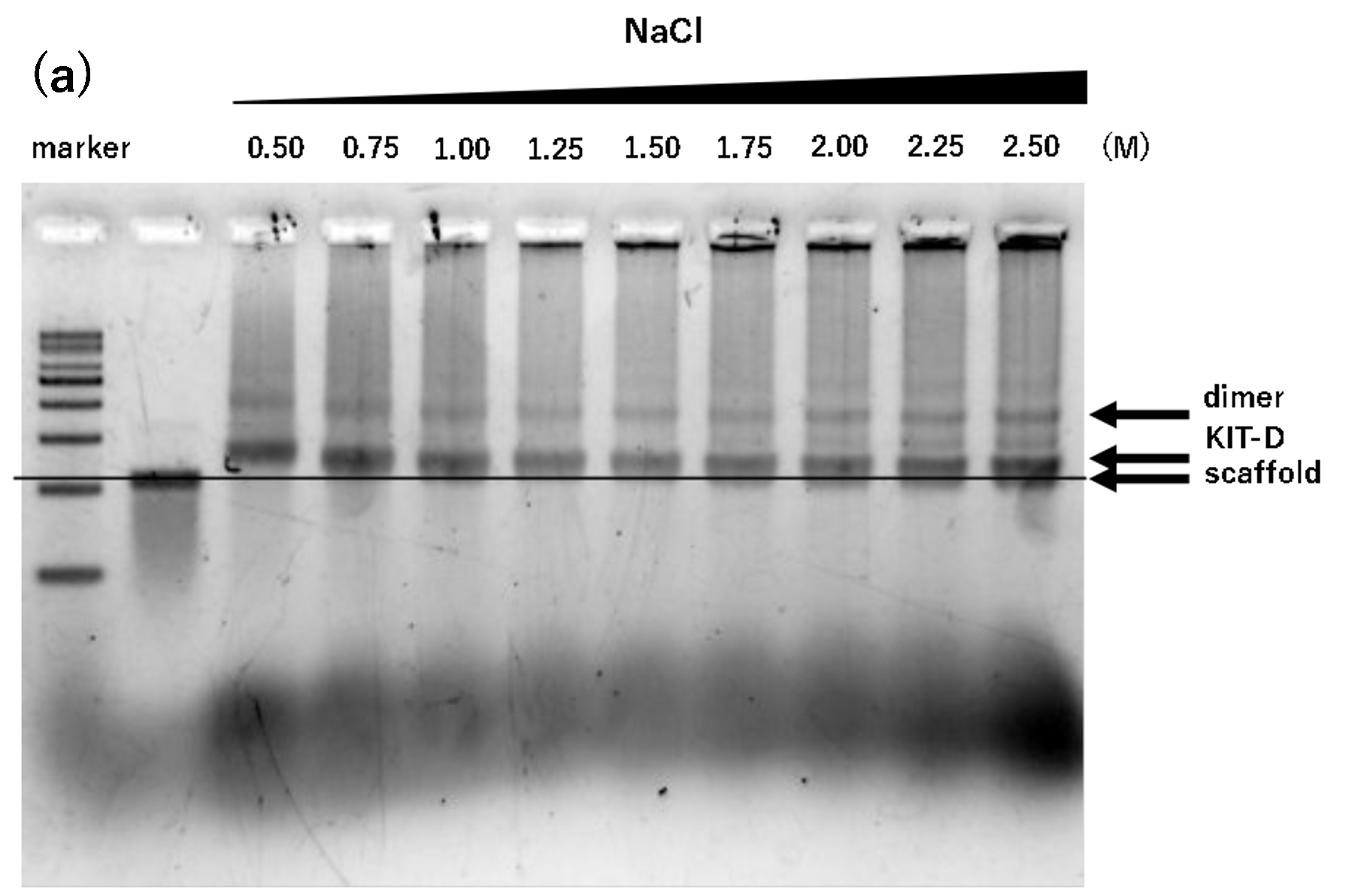
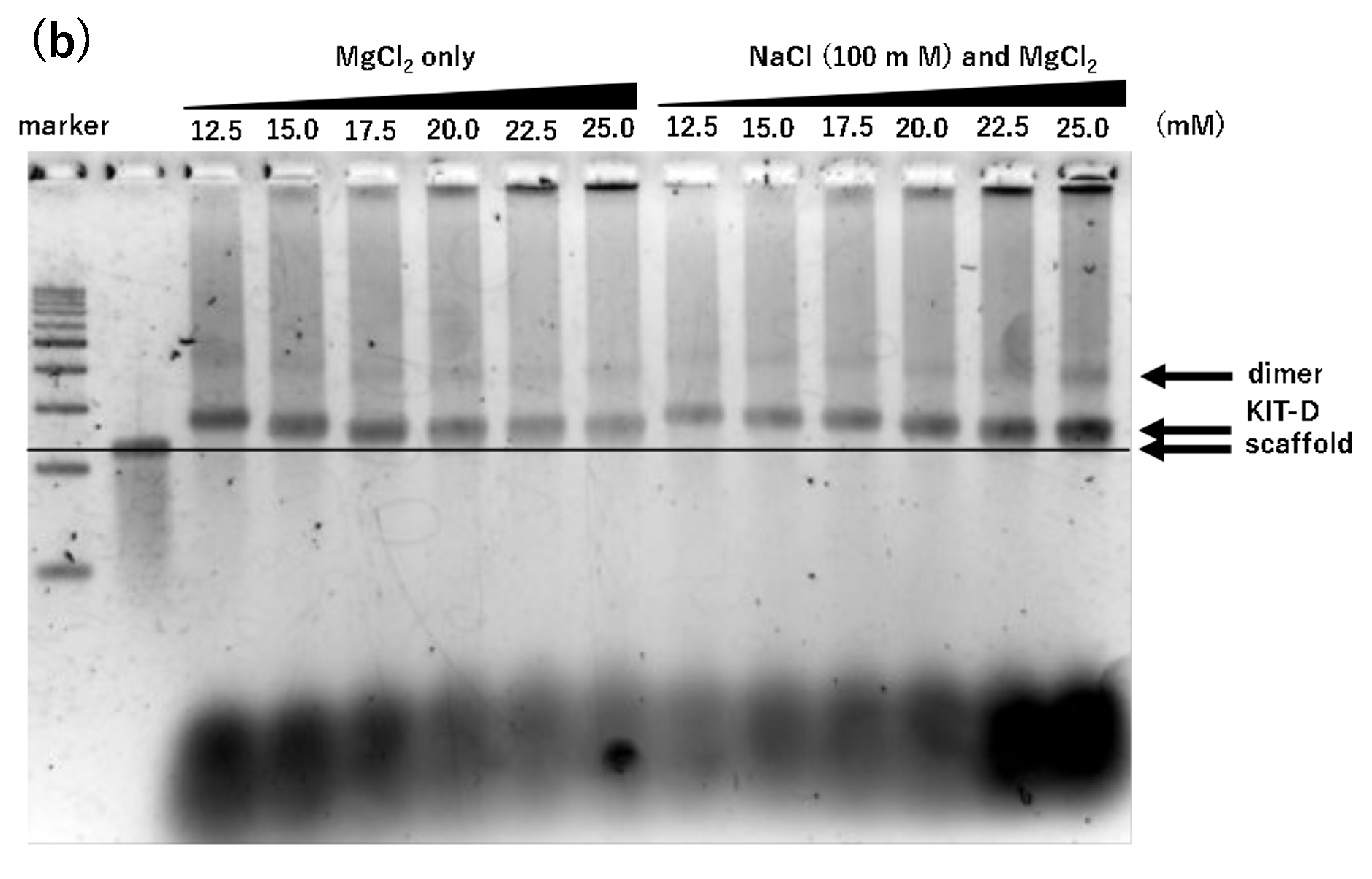
Figure 2. Agarose gel electrophoresis of samples annealed for 17 hours
(a)Na only, 17h, Lane 1: marker, Lane 2: 2 nM scaffold, Lane 3: 0.50
M, Lane 4: 0.75 M, Lane 5: 1.00 M, Lane 6: 1.25 M, Lane 7: 1.50 M,
Lane 8: 1.75 M, Lane 9: 2.00 M, Lane 10: 2.25 M, Lane 11: 2.50 M
(b) Mg only or Na(100 mM) and Mg, 17h, Lane 1: marker, Lane 2: 2 nM
scaffold, Lane 3: 12.5 mM, Lane 4: 15.0 mM, Lane 5: 17.5 mM, Lane 6:
20.0 mM, Lane 7: 22.5 mM, Lane 8: 25.0 mM, Lane 9: 12.5 mM and Na
100 mM, Lane 10: 15.0 mM and Na 100 mM, Lane 11: 17.5 mM and Na 100
mM, Lane 12: 20.0 mM and Na 100 mM, Lane 13: 22.5 mM and Na 100 mM,
Lane 14: 25.0 mM and Na 100 mM