Structure design
The design was performed using the square lattice version on the software caDNAno( https://cadnano.org ). Since KIT-D designed on caDNAno was 7535 bases long, M13mp18 single-strand with a length of 7560 bases was selected as the scaffold strand, and only 25 bases were left over. The total number of staple strands used in the KIT-D construct was 250, both with and without latches, and were designed with the assumption that six identical single-stranded DNA "close" strands would be added to them in order to transform them into the closed form. A file containing the detailed positions and sequences of these designs can be obtained from the following json file.
close_latch9.jsonclose_latch12.json
close_latch15.json
close_latch21.json
file30_withoutlatch.json
KIT-D_sequences.xlsx
Coarse-grained simulation
The structure for simulation was obtained in oxDNA format by converting the json files output from caDNAno using theweb-based tacoxDNA(http://tacoxdna.sissa.it/). The structure was then loaded into theweb-based oxView(https://oxdna.org/static/oxdna-viewer/index.html) and relaxed using rigid body simulations. A two-stage relaxation and molecular dynamics simulation was then performed using a local version of oxDNA. The first stage of the relaxation calculations was a Monte Carlo (MC) simulation using steps, performed at a temperature of 5°C. The second stage was a molecular dynamics (MD) simulation. Temperature 5 deg.Time step (dt) 0.02, 1e6step In the subsequent molecular dynamics simulation, 1e8 steps were performed at a temperature of 25°C and time step (dt) 0.001. The simulation state was saved every 1e6 steps.
Other RMSF
RMSF data for each designed latch length KIT-D structure.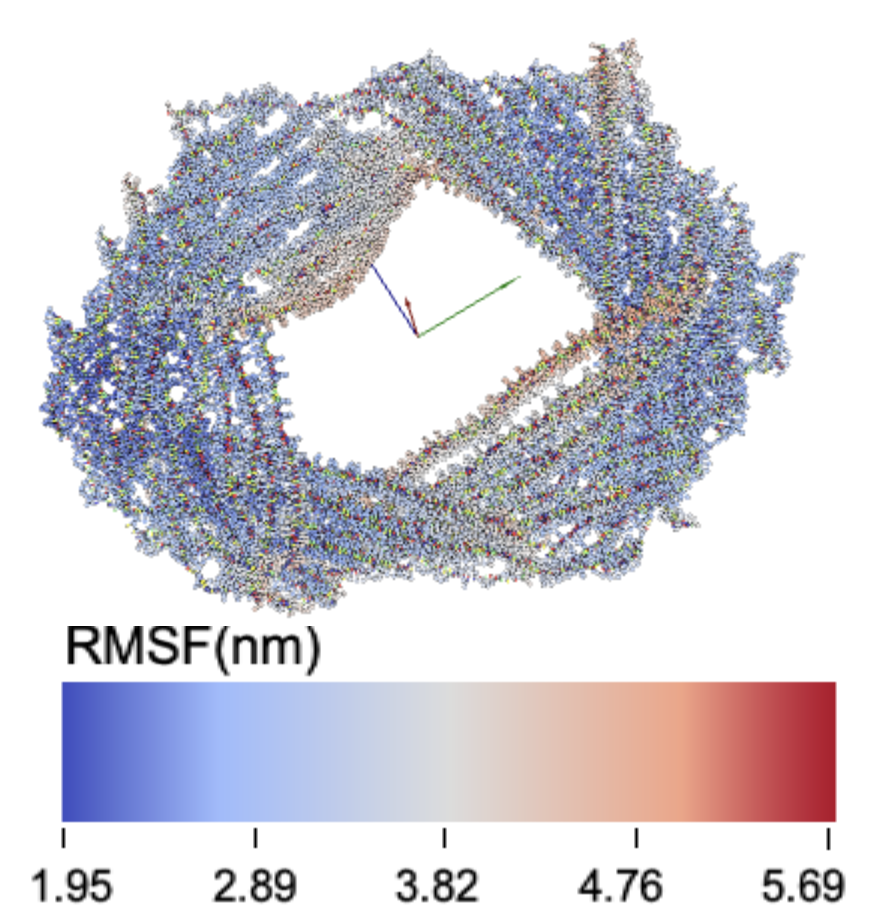
9latch
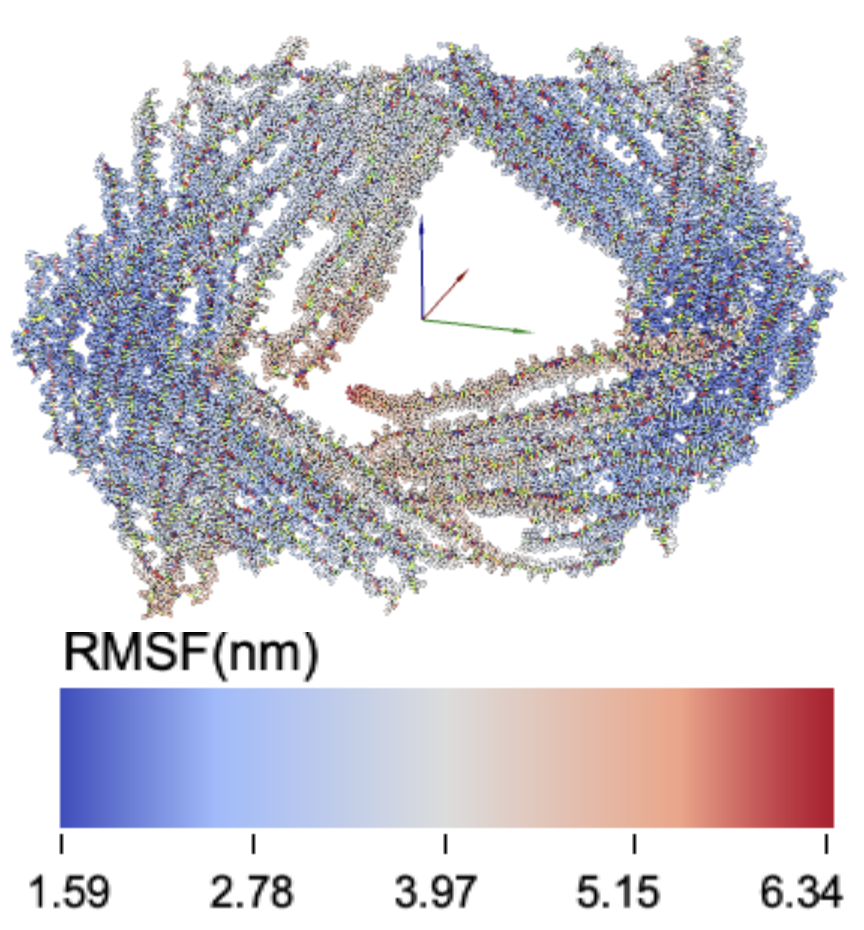
12latch
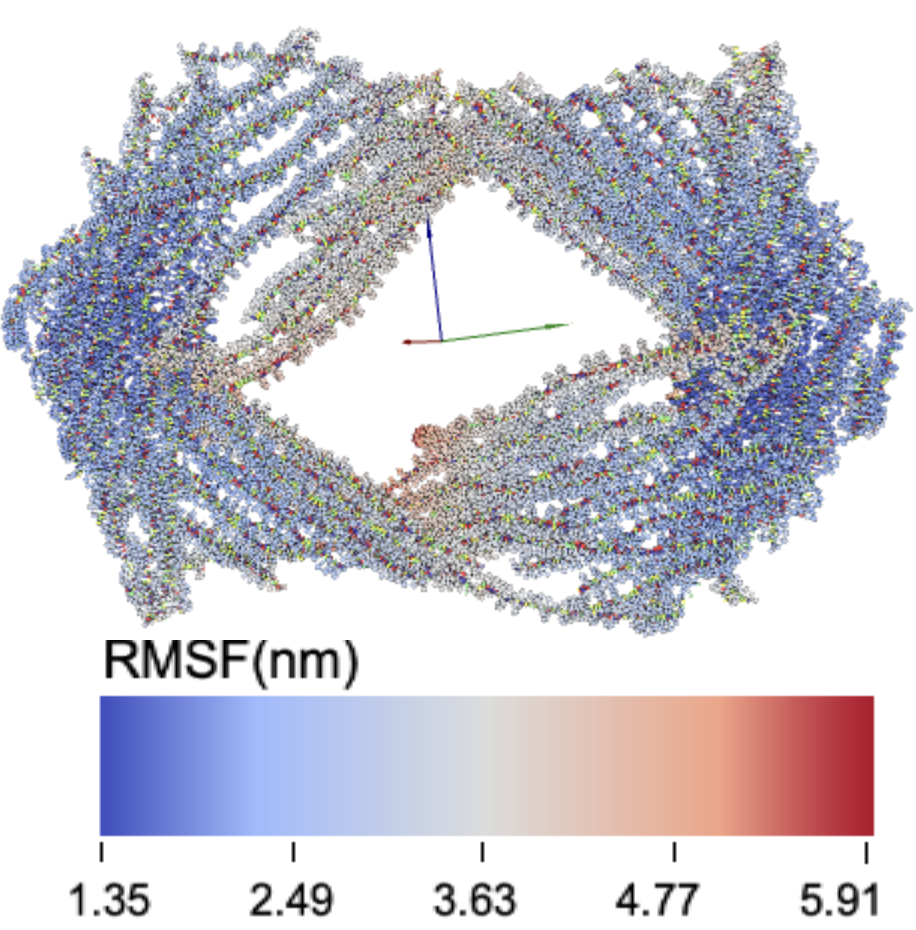
15latch
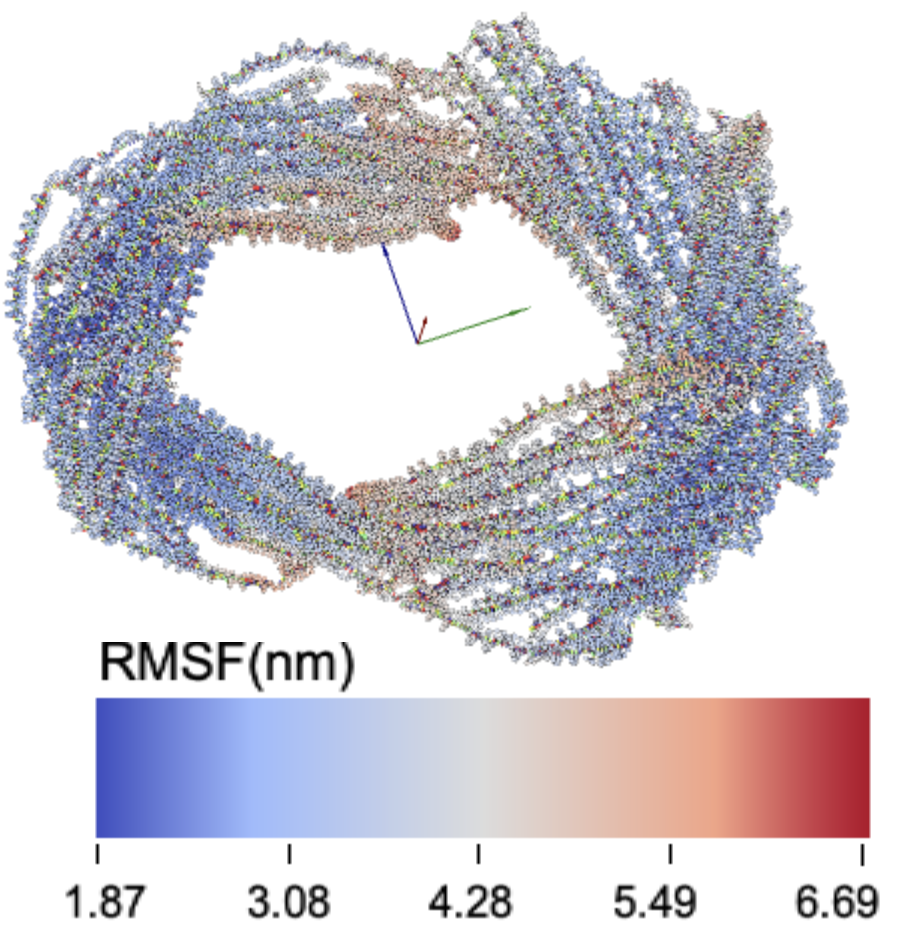
21latch
Preparation of DNA origami structures
Samples were prepared by mixing 15 µL of 400 nM staple strands, 6 µL of 100 nM scaffold strands, 6 μL of 10× folding buffer which consist of 10× TE buffer and ten times the final salt concentration, the rest with MilliQ for a total of 60 μL. For samples with salt concentrations with Na⁺ only, Instead of the 10× folding buffer, 10× TE buffer and the right amount of 5M NaCl solution were added.Annealing was performed by using Biometra TOne thermal cycler, with 60 µL samples aliquoted to three 20 µL samples. During the annealing cycle, the samples were first heated to 85 °C, and then cooled to 65 °C in 5 minutes, then cooled to 60 °C in 15 minutes, 44 °C with -1 °C/hr, and to 4 °C with -1 °C/min. After annealing is complete, the 20 µL samples were combined to one 60 µL sample.
Purification of the samples were done with MicroSpin S-300 HR Columns. Purification gels were set in the columns in advance. First the columns were mixed using a vortex, and then the bottom closure was twisted off and the cap was loosened one-quarter. The column was set onto a 1.5 mL tube and was centrifuged for 800×g for 1 minute. 100 µL of buffer with the same composition as the sample was loaded onto the column and centrifuged at 800×g for 1 minute.repeat this process twice, and then change the 1.5 mL microtube to a new one for retrieval. 50 µL samples were added and were left still for 2 minutes. The output was collected, and its concentration was measured using nanoDrop, and then were compared using agarose gel electrophoresis.
size-exclusion chromatography
https://www.cytivalifesciences.com/en/us/shop/molecular-and-immunodiagnostics/pcr-cleanup-and-size-selection/illustra-microspin-s-300-hr-columns-p-00201?srsltid=AfmBOop_uyKTvATuz49x159aQhJ5j8zPYRf7Nr226Uw-0sIr3QPj9Hcd
Agarose gel electrophoresis analysis
For the agarose gel electrophoresis (AGE) analysis, a 1% agarose gel (KANTO, Japan) was prepared using 0.5X TBE buffer with an additional 5 mM MgCl2. The same buffer solution (0.5X TBE with 5 mM MgCl2) was used as the running buffer. Electrophoresis was conducted at 50 V for 2 hours at 4˚C. A 1 kb DNA Ladder (TaKaRa, Japan) was included as a reference. After electrophoresis, the gel was stained using SYBR Gold (Thermo Fisher Scientific, United States) and visualized with the WSE-6100H LuminoGraph I (ATTO, Japan).Polyacrylamide gel electrophoresis analysis
For polyacrylamide gel electrophoresis (PAGE), the gel was prepared with a composition of 15% (w/v) acrylamide/bis (29:1) solution (Nakalai Tesque, Japan), 1X TBE buffer, and 5 mM MgCl2. The running buffer used was 1X TBE supplemented with 5 mM MgCl2. Electrophoresis was carried out at 100 V for 3 hours at 25˚C. The gel was stained with SYBR Gold (Thermo Fisher Scientific, United States) and visualized using the WSE-6100H LuminoGraph I (ATTO, Japan).TEM imaging
For transmission electron microscopy (TEM) imaging, elastic carbon grids (ELS-C10 STEM Cu100P, Okenshoji, Japan) were used. A 3 μL sample solution was applied to the TEM grid and allowed to incubate for 5 minutes at room temperature. The structures were then stained with 1% (w/v) aqueous uranyl acetate for 5 minutes. After staining, the grids were washed three times with ultrapure water, and excess liquid was removed with filter paper. Imaging was performed using a Tecnai G2 Spirit (FEI, United States) operating at 120 kV.